 The overall tone of the Consumer Genetics Show, held June 2-4, 2010 in Boston MA, was a pragmatic focus on the issues at hand as compared with the enthusiasm and optimism that had marked the conference’s inaugural event last year. One of the biggest shifts was the new programs that some top-tier health service providers have been developing to include genetic testing and interpretation in their organizations. It also became clear that the few widely-agreed upon success stories for genomics in disease diagnosis and drug response (i.e.; warfarin dosing) have been costly to achieve and will not scale to all diseases and all drugs. Cancer continues to be a key killer app for genomics in diagnosis, treatment, prognosis, cancer tumor sequencing, and risk prediction. Appropriate approaches to multigenic risk assessment for health risk and drug response remain untackled. Greater state and federal regulation seems inevitable. Faster-than-Moore’s-law improvements in sequencing costs continue as Illumina dropped the price of whole human genome sequencing for the retail market from $48,000 to $19,500.
The overall tone of the Consumer Genetics Show, held June 2-4, 2010 in Boston MA, was a pragmatic focus on the issues at hand as compared with the enthusiasm and optimism that had marked the conference’s inaugural event last year. One of the biggest shifts was the new programs that some top-tier health service providers have been developing to include genetic testing and interpretation in their organizations. It also became clear that the few widely-agreed upon success stories for genomics in disease diagnosis and drug response (i.e.; warfarin dosing) have been costly to achieve and will not scale to all diseases and all drugs. Cancer continues to be a key killer app for genomics in diagnosis, treatment, prognosis, cancer tumor sequencing, and risk prediction. Appropriate approaches to multigenic risk assessment for health risk and drug response remain untackled. Greater state and federal regulation seems inevitable. Faster-than-Moore’s-law improvements in sequencing costs continue as Illumina dropped the price of whole human genome sequencing for the retail market from $48,000 to $19,500.
There was generally wide agreement that from a public health perspective, personalized genomics is scientifically valid, clinically useful, and reimbursable in specific situations, but not universally – at this time, better information should be obtained for some people, not more information for all people. The focus should be on medical genetics strongly linked to disease, and on pharmacogenomics in treatment. For example, genomic analysis is required for some drugs by the FDA (maraviroc, cetuximab, trastuzumab, and dasatinib), and recommended for several others (warfarin, rasburicase, carbamazepine, abacavir, azathiprine, and irinotecan). A key point is to integrate the drug test with the guidance for drug dosage.
Even when genomic tests are inexpensive enough to be routine, interpretation may be a bottleneck as each individual’s situation is different when taking into account family history, personal medical history, and environmental and other factors. One idea was that the 20,000 pathologists in the US could be a resource for genomic test interpretation; pathologists are already involved as they must certify genetic test data in CLIA labs. Genomic tests and their interpretation would likely need to be standardized and certified in order to be reimbursed in routine medical care. A challenge is that health service payers are not interested or able to drive genomic test product design.
Key science findings
1. The Regulome and Structural Variation
Michael Snyder presented important research that the regulome, the parts of the genome located around the exome (the 1-2% of the genome that codes for protein), may be critical in understanding disease genesis and biological processes. The complexities of RNA are just beginning to be understood. It is known that there is more than just the simple transcription of DNA to RNA involved in controlling gene expression. For example, there is also tight regulation in splicing newly synthesized RNA molecules into the final RNA molecule and in translating messenger RNA to ribosomes to create proteins. Research findings indicate a global/local model of gene regulation, that there are master regulators with universal reach and local regulators operating on a local range of 200 or so genes.
Snyder also presented updates on his lab’s ongoing research into the structural variation of the human genome. A high-resolution sequencing study has been conducted regarding the amount of structural variation in humans, finding that there are ~1,500 structural variations per person that are over 3 kilobases long and that the majority of the structural variations are 3-10 kilobases long with a few extending to 50-100 kilobases (application of this research: Kasowski M, Science, 2010 Apr 9).
2. Reaching beyond the genome to the diseasome, proteome, and microbiome
Several scientists addressed the ways in which science is quickly reaching beyond the single point mutations and structural variation of the genome to other layers of information. There is a need for the digital quantification of the epigenome, the methylome, the transcriptome, the proteome, the metabolome, and the dieaseome/VDJome. For example, the immune system is one of the best monitors of disease state and progression. The strength of individual immune systems can be evaluated through the VDJome (the repertoire of recombined V-D-J regions in immune cells; cumulative immunoglobulin and T-cell receptor antigen exposure)
There are many areas of interest in proteomics including protein profiling, protein-protein interactions, and post-translational modification. A large-scale digital approach to proteomics was presented by Michael Weiner of Affomix. A key focal area is post-translational modification. At least one hundred post-translational modifications have been found, and two are being investigated in particular: phosphorylation (the signal transduction can possibly indicate tumor formation) and glycosylation (possibly indicating tumor progression).
The microbiome (human microbial bacteria) and host-bacteria interactions are an important area for understanding human disease and drug response, and for Procter & Gamble in creating consumer products. The company has basic research and publications underlying products such as the ProX anti-wrinkle skin cream (Hillebrand, Brit Jrl Derm, 2010), rhinovirus, and gingivitis. The company has a substantial vested interest in understanding the microbiome with its variety of nasal, oral, scalp, respiratory, skin, and GI tract-related products.
 DIYgenomics released a personal genome Android app update on January 9, 2011 adding three new elements of functionality: the ability to upload and store 23andMe data in the app, multiple views for health risk, drug response, and athletic performance (Figure 1), and a quality ranking system for each SNP.
DIYgenomics released a personal genome Android app update on January 9, 2011 adding three new elements of functionality: the ability to upload and store 23andMe data in the app, multiple views for health risk, drug response, and athletic performance (Figure 1), and a quality ranking system for each SNP.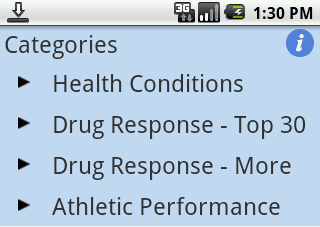















 Email me
Email me Twitter
Twitter MS Futures Group
MS Futures Group Data Visualization Wiki
Data Visualization Wiki Economic Fallacies
Economic Fallacies
